SciPy 2022 virtual poster
by Diptorup Deb and Oleksandr Pavlyk, Intel Corporation
With this poster we would like to inform Scientific Python community about
oneAPI programming model for heterogeneous systems and how to leverage it for
the benefit of Python users.
We hope to interest Python extension authors to start developing portable
accelerator-aware Python packages using oneAPI. This poster presents the tooling
to build Python extensions with DPC++, as well as Python binding to DPC++
runtime classes implemented in dpctl.
1 - Programming model
Get started with oneAPI Python-extensions
In a heterogeneous system there may be multiple devices a Python user may want to engage.
For example, it is common for a consumer-grade laptop to feature an integrated or a discrete
GPU alongside a CPU.
To harness their power one needs to know how to answer the following 3 key questions:
- How does a Python program recognize available computational devices?
- How does a Python workload specify computations to be offloaded to selected devices?
- How does a Python application manage data sharing?
Recognizing available devices
Python package dpctl answers these questions. All the computational devices known to the
underlying DPC++ runtime can be accessed using dpctl.get_devices(). A specific device of
interest can be selected either using a helper function,
e.g. dpctl.select_gpu_device(), or by passing a filter selector string
to dpctl.SyclDevice constructor.
import dpctl
# select a GPU device. If multiple devices present,
# let the underlying runtime select from GPUs
dev_gpu = dpctl.SyclDevice("gpu")
# select a CPU device
dev_cpu = dpctl.SyclDevice("cpu")
# stand-alone function, equivalent to C++
# `auto dev = sycl::gpu_selector().select_device();`
dev_gpu_alt = dpctl.select_gpu_device()
# stand-alone function, equivalent to C++
# `auto dev = sycl::cpu_selector().select_device();`
dev_cpu_alt = dpctl.select_cpu_device()
A device object can be used to query properies of the device, such as its name, vendor, maximal number of computational units, memory size, etc.
Specifying offload target
To answer the second question on the list we need a digression to explain offloading in oneAPI DPC++ first.
Offloading in oneAPI DPC++
In DPC++, a computation kernel can be specified using generic C++ programming and then the kernel can be offloaded to any device that is supported by an underlying SYCL runtime. The device to which the kernel is offloaded is specified using an execution queue when launching the kernel.
The oneAPI unified programming model brings portability across heterogeneous architectures. Another important aspect of the programming model is its inherent flexibility that makes it possible to go beyond portability and even strive for performance portability. An oneAPI library may be implemented using C++ techniques such as template metaprogramming or dynamic polymorphism to implement specializations for a generic kernel. If a kernel is implemented polymorphically, the specialized implementation will be dispatched based on the execution queue specified during kernel launch. The oneMKL library is an example of a performance portable oneAPI library.
A computational task is offloaded for execution on a device by submitting it to DPC++ runtime which inserts the task in a computational graph.
Once the device becomes available the runtime selects a task whose dependencies are met for execution. The computational graph as well as the device targeted by its tasks are stored in a SYCL queue object. The task submission is therefore always
associated with a queue.
Queues can be constructed directly from a device object, or by using a filter selector string to indicate the device to construct:
# construct queue from device object
q1 = dpctl.SyclQueue(dev_gpu)
# construct queue using filter selector
q2 = dpctl.SyclQueue("gpu")
The computational tasks can be stored in an oneAPI native extension in which case their submission is orchestrated
during Python API calls. Let’s consider a function that offloads an evaluation of a polynomial for every point of a
NumPy array X. Such a function needs to receive a queue object to indicate which device the computation must be
offloaded to:
# allocate space for the result
Y = np.empty_like(X)
# evaluate polynomial on the device targeted by the queue, Y[i] = p(X[i])
onapi_ext.offloaded_poly_evaluate(exec_q, X, Y)
Python call to onapi_ext.offloaded_poly_evaluate applied to NumPy arrays of double precision floating pointer numbers gets
translated to the following sample C++ code:
void
cpp_offloaded_poly_evaluate(
sycl::queue q, const double *X, double *Y, size_t n) {
// create buffers from malloc allocations to make data accessible from device
sycl::buffer<1, double> buf_X(X, n);
sycl::buffer<1, double> buf_Y(Y, n);
q.submit([&](sycl::handler &cgh) {
// create buffer accessors indicating kernel data-flow pattern
sycl::accessor acc_X(buf_X, cgh, sycl::read_only);
sycl::accessor acc_Y(buf_Y, cgh, sycl::write_only, sycl::no_init);
cgh.parallel_for(n,
// lambda function that gets executed by different work-items with
// different arguments in parallel
[=](sycl::id<1> id) {
auto x = accX[id];
accY[id] = 3.0 + x * (1.0 + x * (-0.5 + 0.3 * x));
});
}).wait();
return;
}
We refer an interested reader to an excellent and freely available “Data Parallel C++” book for
details of this data parallel C++.
Our package numba_dpex allows one to write kernels directly in Python.
import numba_dpex
@numba_dpex.kernel
def numba_dpex_poly(X, Y):
i = numba_dpex.get_global_id(0)
x = X[i]
Y[i] = 3.0 + x * (1.0 + x * (-0.5 + 0.3 * x))
Specifying the execution queue is done using Python context manager:
import numpy as np
X = np.random.randn(10**6)
Y = np.empty_like(X)
with dpctl.device_context(q):
# apply the kernel to elements of X, writing value into Y,
# while executing using given queue
numba_dpex_poly[X.size, numba_dpex.DEFAULT_LOCAL_SIZE](X, Y)
The argument to device_context can be a queue object, a device object for which a temporary queue will be created,
or a filter selector string. Thus we could have equally used dpctl.device_context(gpu_dev) or dpctl.device_context("gpu").
Note that in this examples data sharing was implicitly managed for us: in the case of calling a function from a precompiled
oneAPI native extension data sharing was managed by DPC++ runtime, while in the case of using numba_dpex kernel it was managed
during execution of __call__ method.
Data sharing
Implicit management of data is surely convenient, but its use in an interpreted code comes at a performance cost. A runtime must
implicitly copy data from host to the device before the kernel execution commences and then copy some (or all) of it back
after the execution completes for every Python API call.
dpctl provides for allocating memory directly accessible to kernels executing on a device using SYCL’s
Unified Shared Memory (USM) feature. It also implements USM-based ND-array
object dpctl.tensor.usm_ndarray that conforms array-API standard.
import dpctl.tensor as dpt
# allocate array of doubles using USM-device allocation on GPU device
X = dpt.arange(0., end=1.0, step=1e-6, device="gpu", usm_type="device")
# allocate array for the output
Y = dpt.empty_like(X)
# execution queue is inferred from allocation queues.
# Kernel is executed on the same device where arrays were allocated
numba_dpex_poly[X.size, numba_dpex.DEFAULT_LOCAL_SIZE](X, Y)
The execution queue can be unambiguously determined in this case since both arguments are
USM arrays with the same allocation queues and X.sycl_queue == Y.sycl_queue evaluates to True.
Should allocation queues be different, such an inference becomes ambiguous and numba_dpex raises
IndeterminateExecutionQueueError advising user to explicitly migrate the data.
Migration can be accomplished either by using dpctl.tensor.asarray(X, device=target_device)
to create a copy, or by using X.to_device(target_device) method.
A USM array can be copied back into a NumPy array using dpt.asnumpy(Y) if needed.
Compute follows data
Automatic deduction of the execution queue from allocation queues is consitent with “local control for data allocation target”
in the array API standard. User has full control over memory allocation through three keyword arguments present in all array creation functions.
For example, consider
# Use usm_type = 'device' to get USM-device allocation (default),
# usm_type = 'shared' to get USM-shared allocation,
# usm_type = 'host' to get USM-host allocation
def dpctl.tensor.empty(..., device=None, usm_type=None, sycl_queue=None) -> dpctl.tensor.usm_ndarray: ...
The keyword device is mandated by the array API. In dpctl.tensor the allowed values of the keyword are
- Filter selector string, e.g.
device="gpu:0" - Existing
dpctl.SyclDevice object, e.g. device=dev_gpu - Existing
dpctl.SyclQueue object dpctl.tensor.Device object instance obtained from an existing USM array, e.g. device=X.device
In all cases, an allocation queue object will be constructed as described earlier and
stored in the array instance, accessible with X.sycl_queue. Instead of using device keyword, one can alternatively
use sycl_queue keyword for readability to directly specify a dpctl.SyclQueue object to be used as the allocation queue.
The rationale for storing the allocation queue in the array is that kernels submitted to this queue are guaranteed to be able to
correctly dereference (i.e. access) the USM pointer. Array operations that only involve this single USM array can thus execute
on the allocation queue, and the output array can be allocated on this same allocation queue with the same usm type as the input array.
Note
Reusing the allocation queue of the input array ensures the computational tasks behind the API call can access the array without making
implicit copies and the output array is allocated on the same device as the input.Compute follows data is the rule prescribing deduction of the execution and the allocation queue as well as the USM type for the result
when multiple USM arrays are combined. It stipulates that arrays can be combined if and only if their allocation queues are the same as measured
by == operator (i.e. X.sycl_queue == Y.sycl_queue must evaluate to True). Same queues refer to the same underlying task graphs and DPC++ schedulers.
An attempt to combine USM arrays with unsame allocation queues raises an exception advising the user to migrate the data.
Migration can be accomplished either by using dpctl.tensor.asarray(X, device=Y.device) to create a copy, or by
using X.to_device(Y.device) method which can sometime do the migration more efficiently.
Advisory
dpctl and numba_dpex are both under heavy development. Feel free to file an issue on GitHub or
reach out on Gitter should you encounter any issues.2 - What is oneAPI
oneAPI - the standard and its implementation.
oneAPI is an open standard for a unified application
programming interface (API) that delivers a common developer experience across
accelerator architectures, including multi-core CPUs, GPUs, and FPGAs.
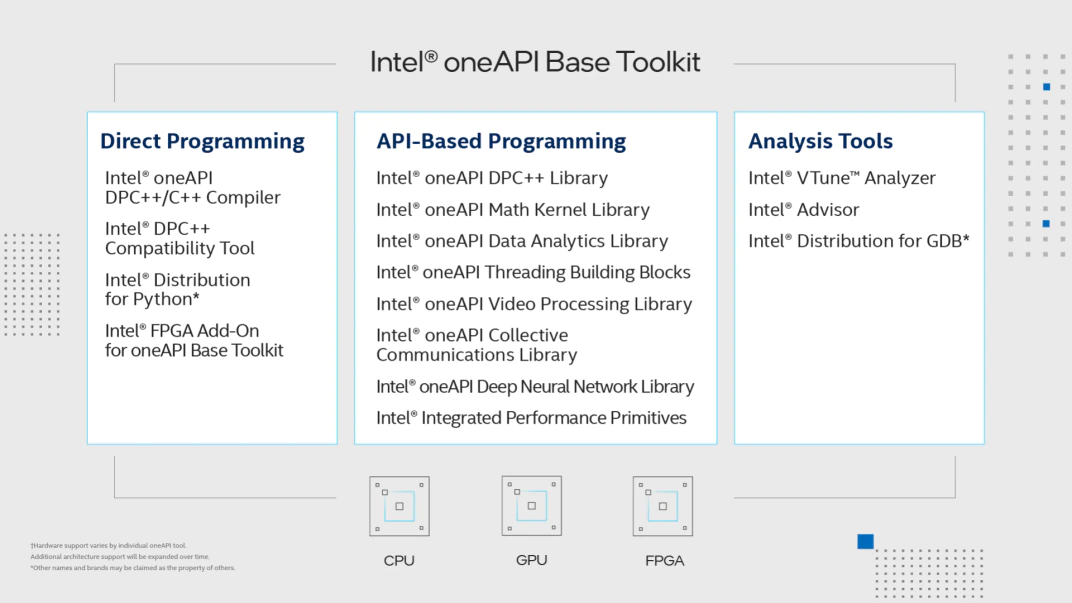
A freely available implementation of the standard is available through
Intel(R) oneAPI Toolkits. The Intel(R) Base Toolkit features
an industry-leading C++ compiler that implements SYCL*, an evolution of C++
for heterogeneous computing. It also includes a suite of performance libraries, such as
Intel(R) oneAPI Math Kernel Library (oneMKL), etc, as well as
Intel(R) Distribution for Python*.
DPC++ is a LLVM-based compiler project that implements compiler and runtime support for SYCL* language.
It is being developed in sycl branch of the LLVM project fork github.com/intel/llvm.
The project publishes daily builds for Linux.
Intel(R) oneAPI DPC++ compiler is a proprietary product that builds on the open-source DPC++ project.
It is part of Intel(R) compiler suite which has completed the adoption of LLVM infrastructure and is available in oneAPI toolkits.
In particular, Intel(R) Fortran compiler is freely avialable on all supported platforms in Intel(R) oneAPI HPC Toolkit.
DPC++ leverages standard toolchain runtime libraries, such as glibc and libstdc++ on Linux and wincrt on Windows. This makes it possible to use
Intel C/C++ compilers, including DPC++, to compile Python native extensions compatible with the CPython and the rest of Python stack.
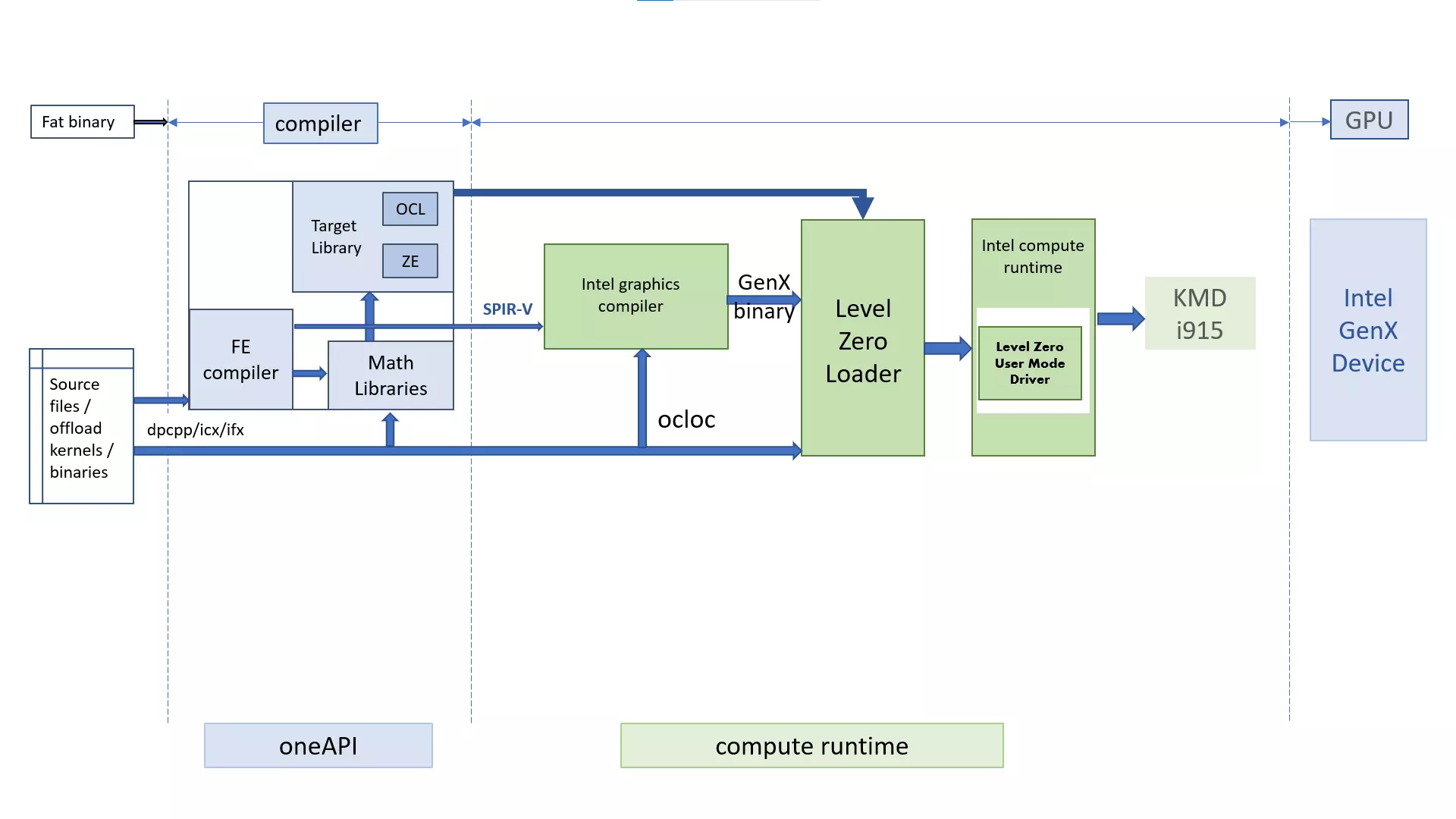
In order to enable cross-architecture programming for CPUs and accelerators the DPC++ runtime adopted layered architecture.
Software concepts are mapped to hardware abstraction layer by user-specified SYCL backend which programs the specific hardware in use.
Compute runtime
An integral part of this layered architecture is provided by Intel(R) Compute Runtime. oneAPI application is a fat binary consisting of
device codes in a standardized intermediate form SPIR-V and host code which orchestrates tasks such as querying of the heterogeneous system it is
running on, selecting accelerator(s), compiling (jitting) device code in the intermediate representation for the selected device, managing device memory, and
submitting compiled device code for execution. The host code performs these tasks by using DPC++ runtime, which maps them to hardware abstraction layer, that
talks to hardware-specific drivers.
Data Parallel C++ book is an excellent resource to get familiar with programming
heterogeneous systems using C++ and SYCL*.
Intel(R) DevCloud hosts base training material which can be executed
on the variety of Intel(R) hardware using preinstalled oneAPI toolkits.
Julia has support for oneAPI github.com/JuliaGPU/oneAPI.jl.
3 - Features Summary
A list of the main features of the data-parallel extensions to Python packages.
DPC++ lets you build cross-platform libraries that can be run on a growing set
of heterogeneous devices supported by the compiler, such as Intel CPUs, GPUs,
and FPGAs, and also Nvidia GPUs and AMD GPUs.
Our package dpctl provides the necessary Python bindings to make a SYCL
library into a Python native extension and subsequently use it from Python.
Write Kernels Directly in Python
If C++ is not your language, you can skip writing data-parallel kernels in SYCL
and directly write them in Python.
Our package numba-dpex extends the Numba compiler to allow kernel
creation directly in Python via a custom compute API.
Cross-architecture Array API
Python array library targeting conformance to core Python Array API
specification.
dpctl.tensor is a Python native extension library implemented using
SYCL within dptcl. The library lets Python users get their job done
using tensor operations powered by pure SYCL generic kernels for portability.
Easy to Install
All the data-parallel extensions for Python packages are readily available for
installation on conda, PyPI, or github.
oneAPI Intel LLVM compilers, including DPC++, as well as associated runtimes are
available on conda to support present and future data-parallel extensions.
4 - Install Intel(R) oneAPI toolkits
Pointers about how to get Intel(R) oneAPI toolkits.
Use Intel(R) DevCloud
Get free access to a development sandbox with preinstalled and configured
oneAPI toolkits as well as access variety of Intel hardware. This is great and low effort way
to start exploring oneAPI. We recommend to start with Jupyter Lab.
Install locally
To add oneAPI to your local toolbox, download and install the basekit for your operating system from download page.
Note
For Linux*, toolkits can be installed using OS’s package managers, as well as tried out from within a docker-container. Please refer
to the download page for specifics.Make sure to configure your system by following steps from “Get Started Guide”
document applicable for your operating system.
Install in CI
oneAPI can be installed into Linux-powered CI by using the OS’s package manager and installing
only the necessary components from required toolkits.
See this example of installing DPC++ compiler in GitHub actions
for IntelPython/dpctl project.
5 - oneAPI Python extensions
Python extensions can be built with DPC++. This is how.
Suitability of DPC++ for Python stack
DPC++ is a single source compiler. It generates both the host code and the device code in a single fat binary.
DPC++ is an LLVM-based compiler, but the host portion of the binary it produces is compatible with GCC runtime libraries on Linux and Windows runtime libraries on Windows. Thus, native Python extensions authored in C++ can be directly built with DPC++. Such extensions will require DPC++ runtime library at the runtime.
Intel(R) compute runtime needs to be present for DPC++ runtime to be able to target supported Intel devices. When using open-source DPC++ from github.com/intel/llvm compiled with support for NVIDIA CUDA, HIP NVIDIA, or HIP AMD (see intel/llvm/getting-started for details), respective runtimes and drivers will need to be present for DPC++ runtime to target these devices.
Build a data-parallel Python native extension
There are two supported ways of building a data-parallel extension: by using
Cython and by using pybind11. The companion repository
IntelPython/sample-data-parallel-extensions provides the
examples demonstrating both approaches by implementing two prototype native
extensions to evaluate Kernel Density Estimate
at a set a points from a Python function with the following signature:
def kde_eval(exec_q: dpctl.SyclQueue, x : np.ndarray, data: np.ndarray, h : float) -> np.narray: ...
"""
Args:
q: execution queue specifying offload target
x: NumPy array of shape (n, dim)
d: NumPy array of shape (n_data, dim)
h: moothing parameter
"""
The examples can be cloned locally using git:
git clone https://github.com/IntelPython/sample-data-parallel-extensions.git
The examples demonstrate a key benefit of using the dpctl package and the
included Cython and pybind11 bindings for oneAPI. By using dpctl,
a native extension writer can focus on writing a data-parallel kernel in DPC++
while automating the generation of the necessary Python bindings using dpctl.
When using setuptools we used environment variables CC and
LDSHARED recognized by setuptools to ensure that dpcpp is used to compile
and link extensions.
CC=dpcpp LDSHARED="dpcpp --shared" python setup.py develop
The resulting extension is a fat binary, containing both the host code with
Python bindings and offloading orchestration, and the device code usually stored
in cross-platform intermediate representation (SPIR-V) and compiled for the
device indicated via the execution queue argument using tooling from compute
runtime.
Building packages with scikit-build
Using setuptools is convenient, but may feel klunky. Using
scikit-build offers an alternate way for users who prefer or are
familiar with CMake.
Scikit-build enables writing the logic of Python package building in CMake which supports oneAPI DPC++. Scikit-build supports building of both
Cython-generated and pybind11-generated native extensions. dpctl integration with CMake allows to conveniently using dpctl integration with these extension generators
simply by including
find_package(Dpctl REQUIRED)
In order for CMake to locate the script that would make the example work, the
example CMakeLists.txt in kde_skbuild package implements
DPCTL_MODULE_PATH variable which can be set to output of python -m dpctl --cmakedir. Integration of DPC++ with CMake requires that CMake’s C and/or C++
compiler were set to Intel LLVM compilers provided in oneAPI base kit.
python setup.py develop -G Ninja -- \
-DCMAKE_C_COMPILER=icx \
-DCMAKE_CXX_COMPILER=icpx \
-DDPCTL_MODULE_PATH=$(python -m dpctl --cmakedir)
Alteratively, we can rely on CMake recognizing CC and CXX environment variables to shorten the input
CC=icx CXX=icpx python setup.py develop -G Ninja -- -DDCPTL_MODULE_PATH=$(python -m dpctl --cmakedir)
Whichever way of building the data-parallel extension appeals to you, the end
result allows offloading computations specified as DPC++ kernels to any
supported device:
import dpctl
import numpy as np
import kde_skbuild as kde
cpu_q = dpctl.SyclQueue("cpu")
gpu_q = dpctl.SyclQueue("gpu")
# output info about targeted devices
cpu_q.print_device_info()
gpu_q.print_device_info()
x = np.linspace(0.1, 0.9, num=14000)
data = np.random.uniform(0, 1, size=10**6)
# Notice that first evaluation results in JIT-compiling the kernel
# Subsequent evaluation reuse cached binary
f0 = kde.cython_kde_eval(cpu_q, x[:, np.newaxis], data[:, np.newaxis], 3e-6)
f1 = kde.cython_kde_eval(gpu_q, x[:, np.newaxis], data[:, np.newaxis], 3e-6)
assert np.allclose(f0, f1)
The following naive NumPy implementation can be used to validate the results
generated by our sample extensions. Do note that the validation script would
not be able to handle very large size inputs and will raise a MemoryError
exception.
def ref_kde(x, data, h):
"""
Reference NumPy implementation for KDE evaluation
"""
assert x.ndim == 2 and data.ndim == 2
assert x.shape[1] == data.shape[1]
dim = x.shape[1]
n_data = data.shape[0]
return np.exp(
np.square(x[:, np.newaxis, :]-data).sum(axis=-1)/(-2*h*h)
).sum(axis=1)/(np.sqrt(2*np.pi)*h)**dim / n_data
Using CPU offload target allows to parallelize CPU computations. For example, try
data = np.random.uniform(0, 1, size=10**3)
x = np.linspace(0.1, 0.9, num=140)
h = 3e-3
%time fr = ref_kde(x[:,np.newaxis], data[:, np.newaxis], h)
%time f0 = kde_skbuild.cython_kde_eval(cpu_q, x[:, np.newaxis], data[:, np.newaxis], h)
%time f1 = kde_skbuild.cython_kde_eval(gpu_q, x[:, np.newaxis], data[:, np.newaxis], h)
assert np.allclose(f0, fr) and np.allclose(f1, fr)
dpctl can be used to build data-parallel Python extensions which functions operating of USM-based arrays.
For example, please refer to examples/pybind11/onemkl_gemv in dpctl sources.
6 - Programming Model
The programming model for the Data Parallel Extensions for Python (DPX4Py)
suite derives from the oneAPI programming model for device
offload. In oneAPI, a computation kernel can be specified using generic C++
programming and then the kernel can be offloaded to any device that is supported
by an underlying SYCL runtime. The device to which the kernel is offloaded is
specified using an execution queue when launching the kernel.
The oneAPI unified programming model brings portability across heterogeneous architectures.
Another important aspect of the programming model is its inherent flexibility
that makes it possible to go beyond portability and even strive for performance
portability. An oneAPI library may be implemented using C++ techniques such as
template metaprogramming or dynamic polymorphism to implement specializations
for a generic kernel. If a kernel is implemented polymorphically, the
specialized implementation will be dispatched based on the execution queue
specified during kernel launch. the oneMKL library is an example of a
performance portable oneAPI library. Figure below shows a gemv kernel from the
oneMKL library that can be launched on multiple types of architectures
simply by changing the execution queue.
TODO: Add gemv figure
In the oneAPI and SYCL programming model, the device where data is allocated is
not tightly coupled with the device where a kernel is executed. The model allows
for implicit data movement across devices. The design offers flexibility to
oneAPI users many of whom are experienced C++ programmers. When extending the
oneAPI programming model to Python via the DPX4Py suite, we felt the need to
make few adjustments to make the model more suited to the Python programming
language. One of the key Python tenets is: explicit is better than
implicit. Following the tenet, a Pythonic programming model for device
offload should allow a programmer to explicitly answer the following two key
questions: Where is data allocated?, Where would the computation occur?
Moreover, if data needs to be moved to a device a programmer should have
explicit control of any such data movement. These requirements are fulfilled by
a programming model called compute follows data.
Compute follows data
TODO:
TODO:
- DPX4Py does support the overall oneAPI programming model. Present current way
of launching kernels in dpex.
- Compute follows data is the prescribed model, but libraries can support
implicit data movement (similar to CuPy or TensorFlow) if the want.
7 - Why use oneAPI in Python
Here is a summary of why we think Scientific Python community should embrace oneAPI
- oneAPI is an open, cross-industry, standards-based, unified, multiarchitecture, multi-vendor programming model.
- DPC++ compiler is being developed in open-source, see http://github.com/intel/llvm, and is being upstreamed into LLVM project itself.
- Open source compiler supports variety of backends, including oneAPI Level-Zero, OpenCL(TM), NVIDIA(R) CUDA(R), and HIP.
- oneAPI Math Kernel Library (oneMKL) Interfaces supports a collection of third-party libraries associated with
supported backends permitting portability.
With these features in mind, and DPC++ runtime being compatible with compiler toolchain used to build CPython itself, use of oneAPI
promises to enable Python extensions to leverage a variety of accelerators, while maintaining portability
of Python extensions across different heterogenous systems, from HPC clusters and servers to laptops.

